Test tutorial
library(dplyr)
## Warning: package 'dplyr' was built under R version 4.1.2
##
## Attaching package: 'dplyr'
## The following objects are masked from 'package:stats':
##
## filter, lag
## The following objects are masked from 'package:base':
##
## intersect, setdiff, setequal, union
grades <- data.frame(
student = c("al", "bo", "cindy", "dan", "ella", "frank", "gina", "henry"),
school = c(rep("stanford", 4), rep("berkley", 4)),
sat_score = c(750, 730, 690, 800, 780, 720, 730, 700)
)
bla bla
grades %>%
group_by(school) %>%
summarize(mean(sat_score))
## # A tibble: 2 × 2
## school `mean(sat_score)`
## <chr> <dbl>
## 1 berkley 732.
## 2 stanford 742.
summary(cars)
## speed dist
## Min. : 4.0 Min. : 2.00
## 1st Qu.:12.0 1st Qu.: 26.00
## Median :15.0 Median : 36.00
## Mean :15.4 Mean : 42.98
## 3rd Qu.:19.0 3rd Qu.: 56.00
## Max. :25.0 Max. :120.00
print(inputFile)
## [1] "/Users/mariokeller/privat/mchicken1988.github.io/tutorials_Rmds/2023-01-07-test_tutorial.Rmd"
library(ggplot2)
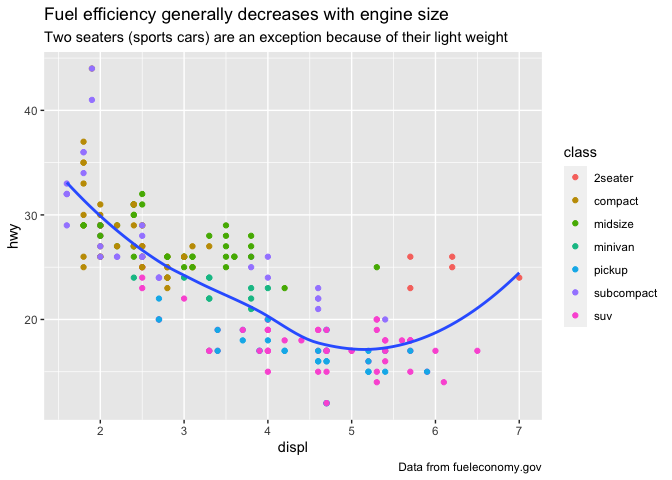
ggplot(mpg, aes(displ, hwy)) +
geom_point(aes(color = class)) +
geom_smooth(se = FALSE, method = "loess") +
labs(
title = "Fuel efficiency generally decreases with engine size",
subtitle = "Two seaters (sports cars) are an exception because of their light weight",
caption = "Data from fueleconomy.gov"
)
## `geom_smooth()` using formula 'y ~ x'
Session Information
sessionInfo()
## R version 4.1.0 (2021-05-18)
## Platform: x86_64-apple-darwin17.0 (64-bit)
## Running under: macOS Big Sur 10.16
##
## Matrix products: default
## BLAS: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRblas.dylib
## LAPACK: /Library/Frameworks/R.framework/Versions/4.1/Resources/lib/libRlapack.dylib
##
## locale:
## [1] en_US.UTF-8/en_US.UTF-8/en_US.UTF-8/C/en_US.UTF-8/en_US.UTF-8
##
## attached base packages:
## [1] stats graphics grDevices utils datasets methods base
##
## other attached packages:
## [1] ggplot2_3.3.5 dplyr_1.0.9
##
## loaded via a namespace (and not attached):
## [1] highr_0.9 compiler_4.1.0 pillar_1.8.0
## [4] GenomeInfoDb_1.29.8 XVector_0.33.0 bitops_1.0-7
## [7] tools_4.1.0 zlibbioc_1.39.0 digest_0.6.28
## [10] lattice_0.20-45 nlme_3.1-153 gtable_0.3.0
## [13] evaluate_0.14 lifecycle_1.0.1 tibble_3.1.8
## [16] mgcv_1.8-37 pkgconfig_2.0.3 rlang_1.0.4
## [19] Matrix_1.3-4 DBI_1.1.1 cli_3.3.0
## [22] rstudioapi_0.13 yaml_2.2.1 xfun_0.26
## [25] fastmap_1.1.0 GenomeInfoDbData_1.2.7 withr_2.4.2
## [28] stringr_1.4.0 knitr_1.36 Biostrings_2.61.2
## [31] generics_0.1.0 S4Vectors_0.31.4 vctrs_0.4.1
## [34] IRanges_2.27.2 grid_4.1.0 stats4_4.1.0
## [37] tidyselect_1.1.1 glue_1.6.2 R6_2.5.1
## [40] fansi_0.5.0 rmarkdown_2.11 farver_2.1.0
## [43] purrr_0.3.4 magrittr_2.0.1 splines_4.1.0
## [46] scales_1.1.1 htmltools_0.5.2 BiocGenerics_0.39.2
## [49] assertthat_0.2.1 colorspace_2.0-2 labeling_0.4.2
## [52] utf8_1.2.2 stringi_1.7.4 munsell_0.5.0
## [55] RCurl_1.98-1.5 crayon_1.5.1